How to restrict the possible parameter range?
There is no built-in way to define hard limits for the parameter and make sure they never go outside of this range.
The recommended way to address this issue is to handle these cases in
the function f you provide.
For example, to keep parameters in the 0-1 range:
p.log.restricted <- function(x) {
if (any(x < 0, x > 1)) {
return(-Inf)
}
B <- 0.03 # controls 'bananacity'
-x[1]^2 / 200 - 1 / 2 * (x[2] + B * x[1]^2 - 100 * B)^2
}
res <- simpleMH(
p.log.restricted,
inits = c(a = 0, b = 0),
theta.cov = diag(2),
max.iter = 3000,
coda = TRUE
)
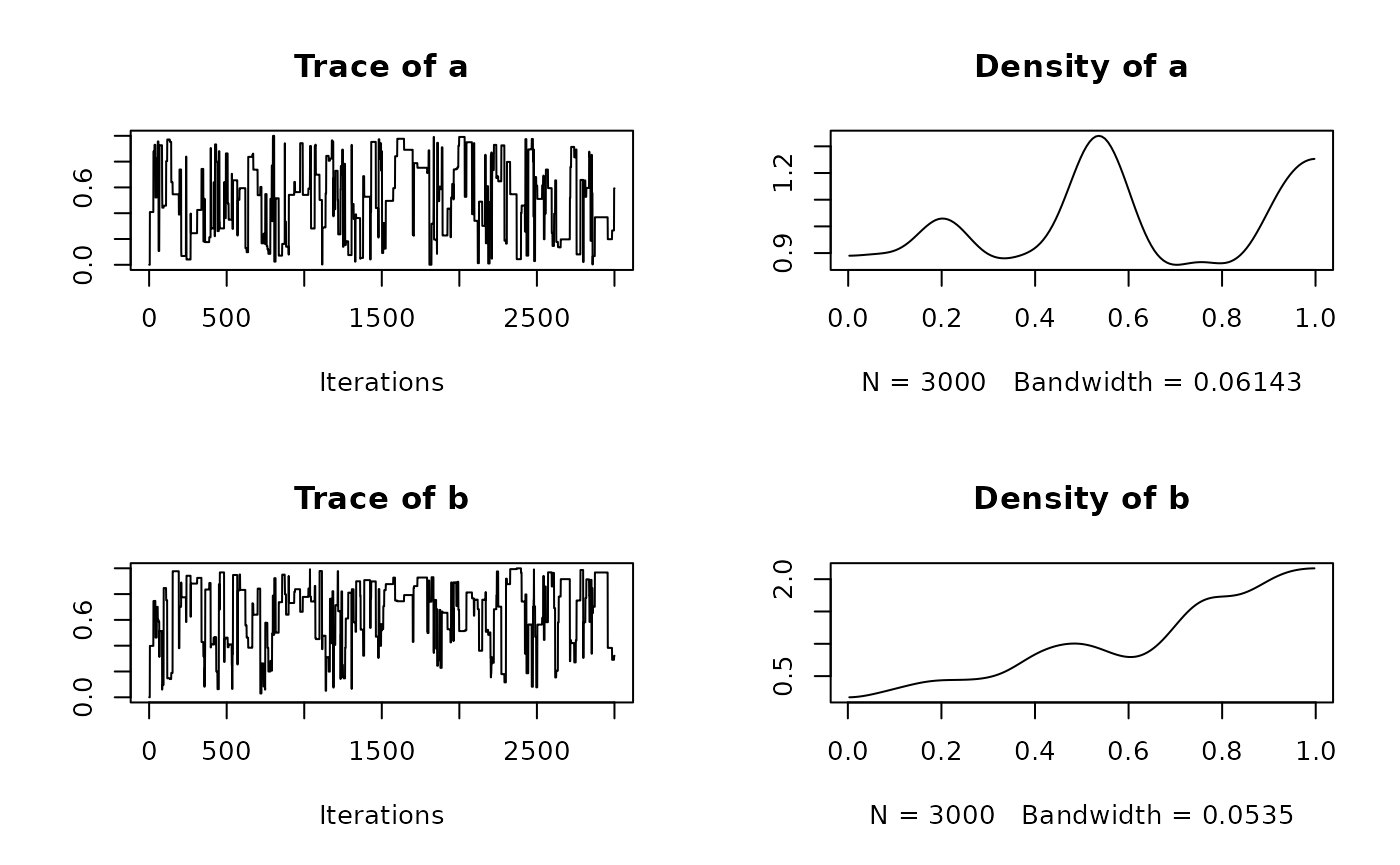
summary(res$samples)
#>
#> Iterations = 1:3000
#> Thinning interval = 1
#> Number of chains = 1
#> Sample size per chain = 3000
#>
#> 1. Empirical mean and standard deviation for each variable,
#> plus standard error of the mean:
#>
#> Mean SD Naive SE Time-series SE
#> a 0.5318 0.2879 0.005256 0.02583
#> b 0.6865 0.2516 0.004594 0.02331
#>
#> 2. Quantiles for each variable:
#>
#> 2.5% 25% 50% 75% 97.5%
#> a 0.05898 0.2603 0.5032 0.7941 0.9781
#> b 0.08251 0.5396 0.7760 0.8816 0.9895
plot(res$samples)